Project
The role of WRKYs in tomato immunity against C. fulvum
This project aims to elucidate how WRKY transcription factors of tomato bridge the upstream and downstream immune signalling pathway. We will determine the role of differentially expressed WRKYs in the tomato defense response by using virus-induced gene silencing (VIGS) and study the function of single WRKYs among the other redundant WRKYs by chimeric repressor silencing technology. We will also identify the proteins that are associated with WRKYs and are required for their function. Furthermore, we aim to verify which downstream defense-related genes that are regulated by WRKYs of tomato.
Introduction
Cladosporium fulvum is a biotrophic fungus, causing tomato (Solanum lycopersicum, Sl) leaf mold disease. The gene-for-gene-based interaction between tomato and C. fulvum is a versatile model to study the resistance mechanism of plants against pathogens and to determine how pathogens circumvent host resistance. Cf resistance genes of tomato mediate the recognition of matching avirulence factors (Avrs) secreted by C. fulvum, which initiates the downstream immune signaling pathway.
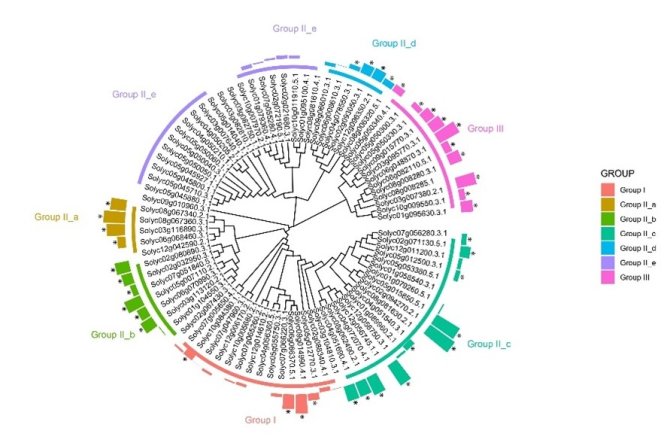
Transcription factors (TFs) are important regulatory proteins with specific DNA-binding domains that bind to DNA enhancer or promotor sequences to either induce or repress the transcription of genes, thereby allowing plants to adapt to a changing environment. The WRKY family is very large and the best-characterized plant-specific class of TFs, of which the members generally contain one or two 60-amino acids, four-stranded β-sheet WRKY DNA binding domains (DBD), having the conserved core sequence WRKYGQK at their N-terminus and a zinc finger motif at their C-terminus. Most WRKY TFs are known to regulate target gene expression by binding to their promoter region, which includes the so-called W box, which has the conserved sequence TTGACC/T. For now, 83 SlWRKY genes have been identified in tomato, which are distributed over all 12 chromosomes, except for chromosome 11. SlWRKY proteins are the most important TFs in regulating transcriptional reprogramming upon the initiation of immune signaling leading to the hypersensitive response (HR). SlWRKYs impact plant tolerance to both biotic and abiotic stresses, both positively and negatively.
Project description
The WRKY family of TFs plays a crucial role in various biological processes in plants, including the response to abiotic stress, development, and defense against pathogens. Although research focuses on the structure of the various WRKYs, their classification, and function, most of these studies are being performed in model plants, like Arabidopsis thaliana and rice. Despite the importance of WRKY TFs, several major knowledge gaps in this field still need to be addressed. First, we will determine the specific functions of SlWRKY TFs in the tomato-C. fulvum interaction and will identify the mechanisms by which they contribute to defense against the invading fungus, for example by using VIGS of individual WRKYs or WRKY families. Then, we will study the function of single SlWRKYs among other redundant SlWRKYs and decipher how they contribute to the overall function of the WRKY family by chimeric repressor silencing technology. Moreover, advanced technologies aimed at deciphering the composition of protein complexes, such as proximity-dependent labelling using the TurboID biotinylating enzyme, will be used to identify the proteins interacting with SlWRKYs to regulate their function.
Results